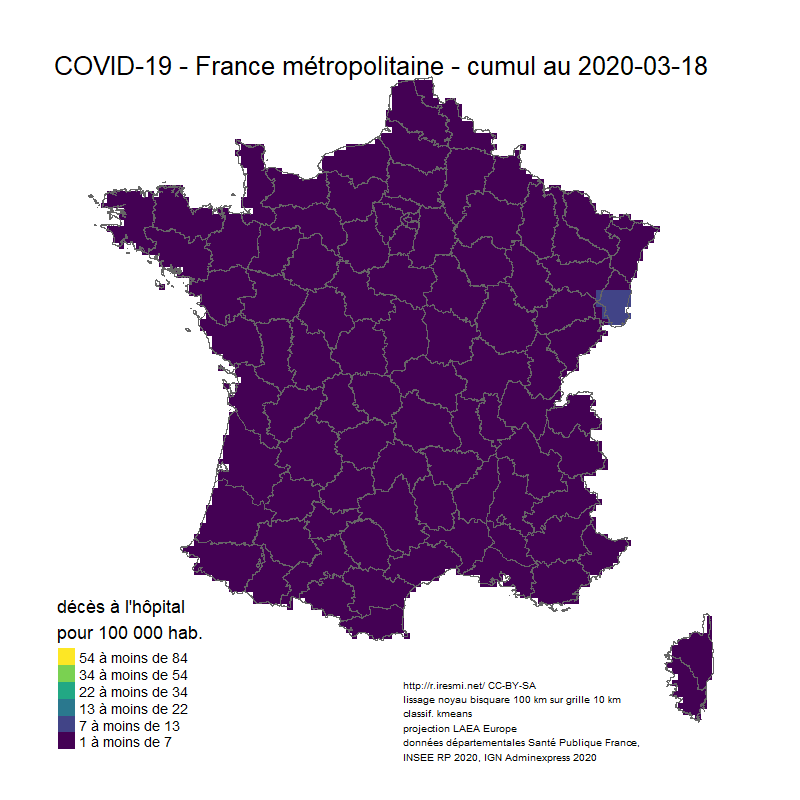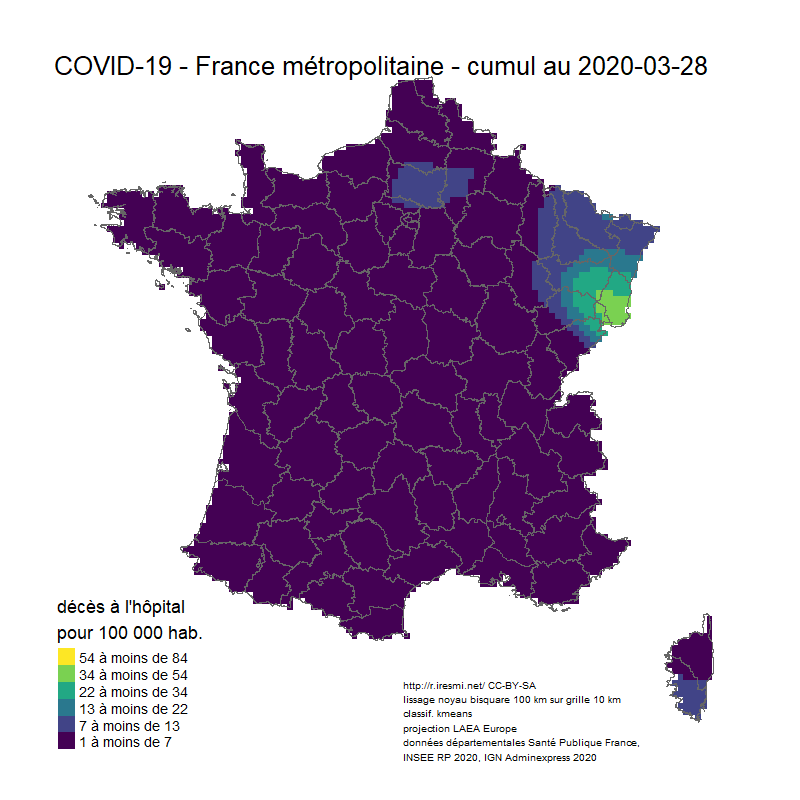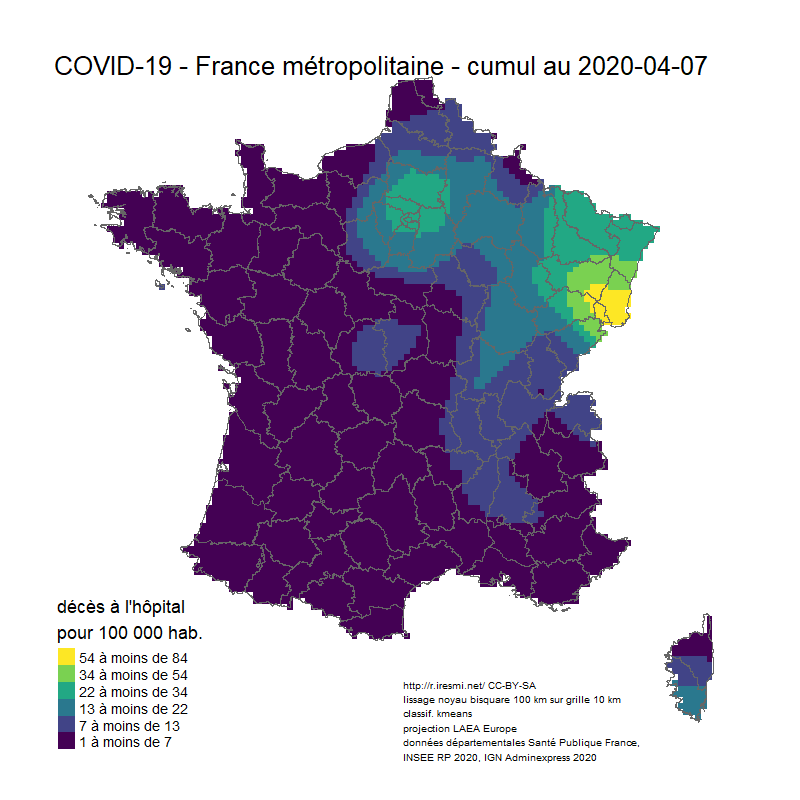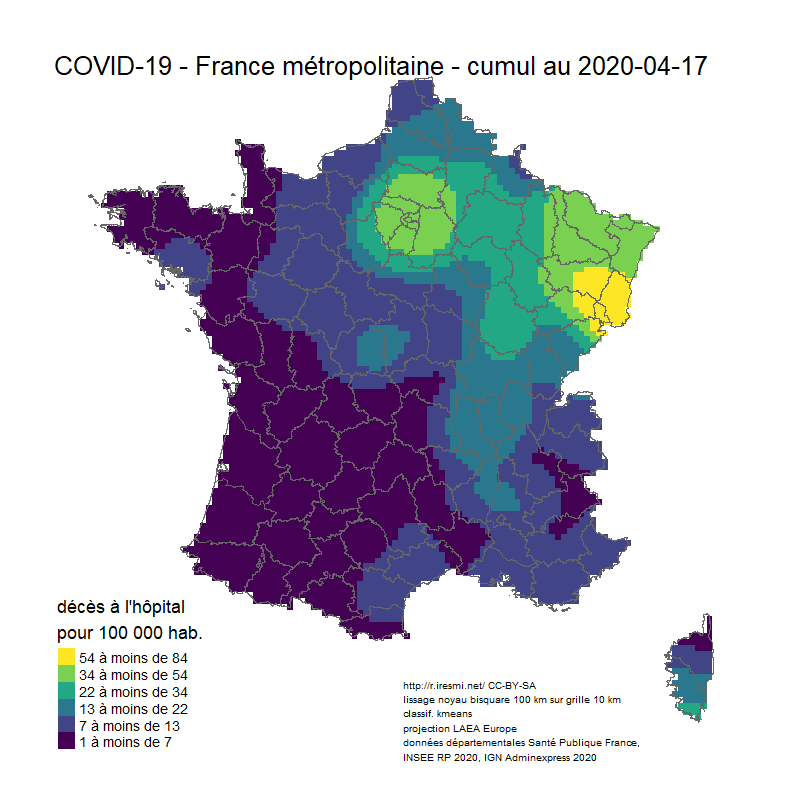
# Animation carto décès COVID 19 France
# avec lissage
# packages -----------------------------------------------------------------
library(tidyverse)
library(httr)
library(fs)
library(sf)
library(readxl)
library(janitor)
library(glue)
library(tmap)
library(grid)
library(classInt)
library(magick)
# + btb, raster, fasterize, plyr
# sources -----------------------------------------------------------------
# https://www.data.gouv.fr/fr/datasets/donnees-hospitalieres-relatives-a-lepidemie-de-covid-19/
fichier_covid <- "donnees/covid.csv"
url_donnees_covid <- "https://www.data.gouv.fr/fr/datasets/r/63352e38-d353-4b54-bfd1-f1b3ee1cabd7"
# https://www.insee.fr/fr/statistiques/2012713#tableau-TCRD_004_tab1_departements
fichier_pop <- "donnees/pop.xlsx"
url_donnees_pop <- "https://www.insee.fr/fr/statistiques/fichier/2012713/TCRD_004.xlsx"
# Adminexpress : à télécharger manuellement
# https://geoservices.ign.fr/documentation/diffusion/telechargement-donnees-libres.html#admin-express
aex <- path_expand("~/data/adminexpress/adminexpress_cog_simpl_000_2022.gpkg")
# config ------------------------------------------------------------------
rayon <- 100000 # distance de lissage (m)
pixel <- 10000 # résolution grille (m)
force_download <- FALSE # retélécharger même si le fichier existe et a été téléchargé aujourd'hui ?
#' Kernel weighted smoothing with arbitrary bounding area
#'
#' @param df sf object (points)
#' @param field weight field in the df
#' @param bandwidth kernel bandwidth (map units)
#' @param resolution output grid resolution (map units)
#' @param zone sf study zone (polygon)
#' @param out_crs EPSG (should be an equal-area projection)
#'
#' @return a raster object
#' @import btb, raster, fasterize, dplyr, plyr, sf
lissage <- function(df, field, bandwidth, resolution, zone, out_crs = 3035) {
if (st_crs(zone)$epsg != out_crs) {
message("reprojecting data...")
zone <- st_transform(zone, out_crs)
}
if (st_crs(df)$epsg != out_crs) {
message("reprojecting study zone...")
df <- st_transform(df, out_crs)
}
zone_bbox <- st_bbox(zone)
# grid generation
message("generating reference grid...")
zone_xy <- zone %>%
dplyr::select(geometry) %>%
st_make_grid(
cellsize = resolution,
offset = c(
plyr::round_any(zone_bbox[1] - bandwidth, resolution, f = floor),
plyr::round_any(zone_bbox[2] - bandwidth, resolution, f = floor)
),
what = "centers"
) %>%
st_sf() %>%
st_join(zone, join = st_intersects, left = FALSE) %>%
st_coordinates() %>%
as_tibble() %>%
dplyr::select(x = X, y = Y)
# kernel
message("computing kernel...")
kernel <- df %>%
cbind(., st_coordinates(.)) %>%
st_set_geometry(NULL) %>%
dplyr::select(x = X, y = Y, field) %>%
btb::btb_smooth(
pts = .,
sEPSG = out_crs,
iCellSize = resolution,
iBandwidth = bandwidth,
vQuantiles = NULL,
dfCentroids = zone_xy
)
# rasterization
message("\nrasterizing...")
raster::raster(
xmn = plyr::round_any(zone_bbox[1] - bandwidth, resolution, f = floor),
ymn = plyr::round_any(zone_bbox[2] - bandwidth, resolution, f = floor),
xmx = plyr::round_any(zone_bbox[3] + bandwidth, resolution, f = ceiling),
ymx = plyr::round_any(zone_bbox[4] + bandwidth, resolution, f = ceiling),
resolution = resolution
) %>%
fasterize::fasterize(kernel, ., field = field)
}
# téléchargement--------------------------------------------------------------
dir_create("donnees")
dir_create("resultats")
dir_create("resultats/animation")
if (!file_exists(fichier_covid) |
file_info(fichier_covid)$modification_time < Sys.Date() |
force_download) {
GET(url_donnees_covid,
progress(),
write_disk(fichier_covid, overwrite = TRUE))
}
if (!file_exists(fichier_pop)) {
GET(url_donnees_pop,
progress(),
write_disk(fichier_pop))
}
# données -----------------------------------------------------------------
covid <- read_csv2(fichier_covid) %>%
filter(jour < "2020-05-01")
# adminexpress prétéléchargé
dep <- read_sf(aex, layer = "departement") %>%
filter(insee_dep < "971")
pop <- read_xlsx(fichier_pop, skip = 2) %>%
clean_names() %>%
select(insee_dep = x1,
x2020)
# prétraitement -----------------------------------------------------------
# contour métropole pour grille de référence
fichier_fr <- "donnees/fr.rds"
if (!file_exists(fichier_fr)) {
fr <- dep %>%
st_union() %>%
st_sf() %>%
write_rds(fichier_fr)
} else {
fr <- read_rds(fichier_fr)
}
# jointures des données
creer_df <- function(territoire, date = NULL) {
territoire %>%
left_join(pop, join_by(insee_dep)) %>%
left_join(
covid %>%
filter(jour == ifelse(is.null(date), max(jour), date),
sexe == 0) %>%
rename(deces = dc,
reanim = rea,
hospit = hosp),
join_by(insee_dep == dep)) %>%
st_point_on_surface()
}
covid_geo_pop <- creer_df(dep)
# lissage -----------------------------------------------------------------
# génération de la dernière grille mortalité
# et création des grilles pour 100000 habitants
# décès métropole
d <- covid_geo_pop %>%
lissage("deces", rayon, pixel, fr)
# population métropole et DOM
p <- covid_geo_pop %>%
lissage("x2020", rayon, pixel, fr)
# grilles pour 100000 hab
d100k <- d * 1e5 / p
# classification à réutiliser pour les autres cartes
set.seed(1234)
classes <- classIntervals(raster::values(d100k), n = 6, style = "kmeans", dataPrecision = 0)$brks
# animation ---------------------------------------------------------------
image_animation <- function(date) {
m <- creer_df(dep, date) %>%
lissage("deces", rayon, pixel, fr) %>%
magrittr::divide_by(p) %>%
magrittr::multiply_by(100000) %>%
tm_shape() +
tm_raster(title = glue("décès à l'hôpital
pour 100 000 hab."),
style = "fixed",
breaks = classes,
palette = "viridis",
legend.format = list(text.separator = "à moins de",
digits = 0),
legend.reverse = TRUE) +
tm_shape(dep) +
tm_borders() +
tm_layout(title = glue("COVID-19 - France métropolitaine - cumul au {date}"),
legend.position = c("left", "bottom"),
frame = FALSE) +
tm_credits(glue("http://r.iresmi.net/
lissage noyau bisquare {rayon / 1000} km sur grille {pixel / 1000} km
classif. kmeans
projection LAEA Europe
données départementales Santé publique France,
INSEE RP 2020, IGN Adminexpress 2020"),
size = .5,
position = c(.5, .025))
tmap_save(m, glue("resultats/animation/covid_fr_{date}.png"),
width = 800, height = 800, scale = .4,)
}
unique(covid$jour) %>%
walk(image_animation)
animation <- glue("resultats/deces_covid19_fr_{max(covid$jour)}.gif")
dir_ls("resultats/animation") %>%
map(image_read) %>%
image_join() %>%
#image_scale("500x500") %>%
image_morph(frames = 1) %>%
image_animate(fps = 2, optimize = TRUE) %>%
image_write(animation)