# Register an account on https://www.gbif.org
# and save the credentials in ~/.Renviron :
# GBIF_USER="xxxx"
# GBIF_PWD="*****"
# GBIF_EMAIL="you@example.org"
library(rgbif)
library(tidyverse)
library(sf)
library(rnaturalearth)
library(ggtext)
library(glue)
# basemap
countries <- ne_countries(scale = "medium", returnclass = "sf")
# Get ID of the species
taxon <- name_backbone("Lynx lynx")Day 1 of 30DayMapChallenge: « Points » (previously).
Using data from GBIF to map species presence points.
# Launch the query
occ_download(pred("taxonKey", taxon$usageKey),
format = "SIMPLE_CSV")# Query status (your ID will differ)
occ_download_wait("0034730-231002084531237")<<gbif download metadata>>
Status: SUCCEEDED
DOI: 10.15468/dl.jfaebj
Format: SIMPLE_CSV
Download key: 0034730-231002084531237
Created: 2023-11-01T17:52:20.752+00:00
Modified: 2023-11-01T17:54:02.517+00:00
Download link: https://api.gbif.org/v1/occurrence/download/request/0034730-231002084531237.zip
Total records: 77187# Get results
lynx <- occ_download_get("0034730-231002084531237") |>
occ_download_import()
glimpse(lynx)Rows: 77,187
Columns: 50
$ gbifID <int64> 991907429, 924537973, 924537972, 92…
$ datasetKey <chr> "b7ec1bf8-819b-11e2-bad2-00145eb45e9a…
$ occurrenceID <chr> "", "7408f0e4-f81d-11e6-ae10-70e2840f…
$ kingdom <chr> "Animalia", "Animalia", "Animalia", "…
$ phylum <chr> "Chordata", "Chordata", "Chordata", "…
$ class <chr> "Mammalia", "Mammalia", "Mammalia", "…
$ order <chr> "Carnivora", "Carnivora", "Carnivora"…
$ family <chr> "Felidae", "Felidae", "Felidae", "Fel…
$ genus <chr> "Lynx", "Lynx", "Lynx", "Lynx", "Lynx…
$ species <chr> "Lynx lynx", "Lynx lynx", "Lynx lynx"…
$ infraspecificEpithet <chr> "", "", "", "", "", "", "", "", "", "…
$ taxonRank <chr> "SPECIES", "SPECIES", "SPECIES", "SPE…
$ scientificName <chr> "Lynx lynx (Linnaeus, 1758)", "Lynx l…
$ verbatimScientificName <chr> "Lynx lynx Linnaeus, 1758", "Lynx lyn…
$ verbatimScientificNameAuthorship <chr> "", "", "", "", "", "", "", "", "", "…
$ countryCode <chr> "DE", "", "", "SE", "DE", "CA", "DE",…
$ locality <chr> "Tierpark Berlin", "", "", "Vansbro K…
$ stateProvince <chr> "", "", "", "", "", "", "", "", "", "…
$ occurrenceStatus <chr> "PRESENT", "PRESENT", "PRESENT", "PRE…
$ individualCount <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ publishingOrgKey <chr> "10980920-6dad-11da-ad13-b8a03c50a862…
$ decimalLatitude <dbl> NA, NA, NA, 60.52402, 51.63549, 50.80…
$ decimalLongitude <dbl> NA, NA, NA, 14.179573, 10.787888, -10…
$ coordinateUncertaintyInMeters <dbl> NA, NA, NA, 250, 250, 250, 250, 250, …
$ coordinatePrecision <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ elevation <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ elevationAccuracy <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ depth <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ depthAccuracy <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ eventDate <dttm> 1962-01-13 15:00:00, NA, NA, 2003-07…
$ day <int> 13, NA, NA, 24, 15, 28, 6, 6, 28, 6, …
$ month <int> 1, NA, NA, 7, 3, 8, 5, 8, 5, 8, 1, 9,…
$ year <int> 1962, NA, NA, 2003, 2012, 2011, 2011,…
$ taxonKey <int> 2435240, 2435240, 2435240, 2435240, 2…
$ speciesKey <int> 2435240, 2435240, 2435240, 2435240, 2…
$ basisOfRecord <chr> "OBSERVATION", "PRESERVED_SPECIMEN", …
$ institutionCode <chr> "MfN", "KPM", "KPM", "naturgucker", "…
$ collectionCode <chr> "TSA", "NFM", "NFM", "naturgucker", "…
$ catalogNumber <chr> "TSA:Lynx_lynx_S_674_2_1", "KPM-NFM00…
$ recordNumber <chr> "", "", "", "", "", "", "", "", "", "…
$ identifiedBy <chr> "", "", "", "", "", "", "", "", "", "…
$ dateIdentified <dttm> NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ license <chr> "CC_BY_4_0", "CC_BY_4_0", "CC_BY_4_0"…
$ rightsHolder <chr> "", "", "", "", "", "", "", "", "", "…
$ recordedBy <chr> "Tembrock, Günter", "", "", "34172526…
$ typeStatus <chr> "", "", "", "", "", "", "", "", "", "…
$ establishmentMeans <chr> "", "", "", "", "", "", "", "", "", "…
$ lastInterpreted <dttm> 2023-08-26 10:10:45, 2023-10-14 04:4…
$ mediaType <chr> "Sound", "", "", "", "", "", "", "", …
$ issue <chr> "CONTINENT_DERIVED_FROM_COUNTRY", "IN…# Some cleaning (valid coords, presence, on land)
lynx_geo <- lynx |>
drop_na(decimalLongitude, decimalLatitude) |>
filter(occurrenceStatus == "PRESENT",
!near(decimalLongitude, 0) & !near(decimalLatitude, 0)) |>
st_as_sf(coords = c("decimalLongitude", "decimalLatitude"), crs = "EPSG:4326")# Map
ggplot(lynx_geo) +
geom_sf(data = countries) +
geom_sf(color = "red", size = 0.1, alpha = 0.05) +
coord_sf(xlim = c(2800000, 7000000), ylim = c(2000000, 5300000), crs = "EPSG:3035") +
labs(title = "_Lynx lynx_ occurrence",
subtitle = glue("Europe, {min(lynx_geo$year, na.rm = TRUE)} - {max(lynx_geo$year, na.rm = TRUE)}"),
caption = glue("GBIF occurrence download https://doi.org/10.15468/dl.jfaebj
Basemap: Natural Earth
https://r.iresmi.net/")) +
theme(plot.caption = element_text(size = 7),
plot.title = element_markdown())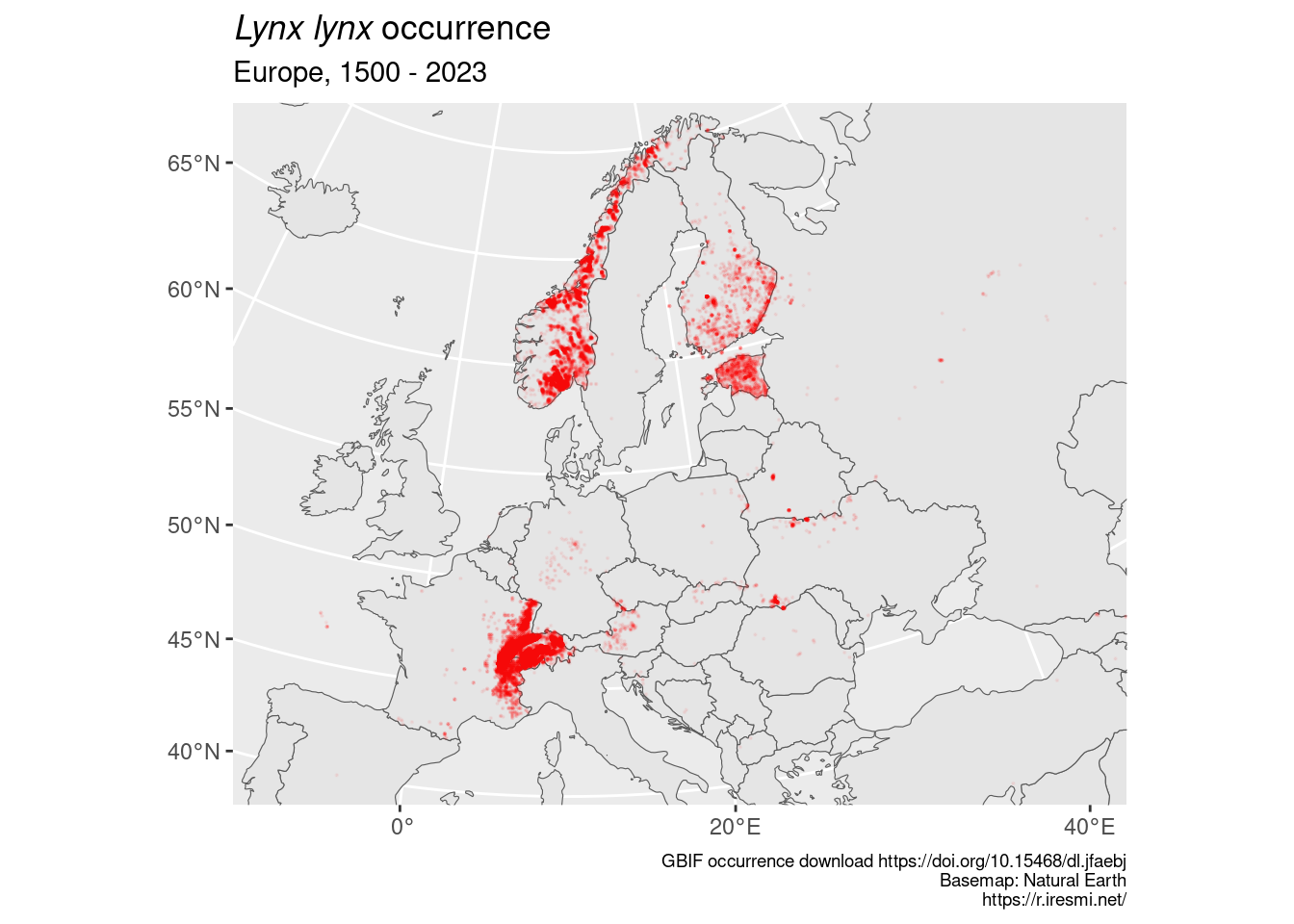
Well, I guess the contributions are unequal between countries (Sweden?) and some cleaning should be done (sightings in North Atlantic and at Null Island!).
There is a lot of overplotting; we should do some smoothing…
