Codes postaux
R
spatial
datavisualization
30DayMapChallenge
french
Bike accidents
R
spatial
datavisualization
30DayMapChallenge
OSM
Greenland ice thickness
R
spatial
datavisualization
30DayMapChallenge
raster
Birth date temperature
R
spatial
datavisualization
30DayMapChallenge
raster
Whales movements
R
spatial
datavisualization
30DayMapChallenge
ecology
Global discret grids
R
spatial
datavisualization
30DayMapChallenge
Street names
R
spatial
datavisualization
Parquet
OSM
30DayMapChallenge
Birds in atmosphere
R
30DayMapChallenge
spatial
datavisualization
ecology
El niño anomaly
R
30DayMapChallenge
spatial
datavisualization
remote sensing
Defibrillator from OSM
R
30DayMapChallenge
spatial
datavisualization
OSM
Renewable energy in Europe
R
30DayMapChallenge
spatial
datavisualization
Clustering points to polygons
R
30DayMapChallenge
spatial
datavisualization
OSM Guadeloupe trail relation
R
30DayMapChallenge
spatial
OSM
datavisualization
Map your Strava activities
R
datavisualization
spatial
sport
Sentinel
R
30DayMapChallenge
datavisualization
spatial
raster
remote sensing
Use data from Wikipedia
R
30DayMapChallenge
datavisualization
spatial
webscraping
Wikipedia
The giant French Olympic-size swimming pool
R
30DayMapChallenge
datavisualization
spatial
OSM
Use data from Openstreetmap
R
30DayMapChallenge
datavisualization
spatial
OSM
Opening a spatial subset with {sf}
R
30DayMapChallenge
datavisualization
spatial
My air travel carbon footprint
R
30DayMapChallenge
datavisualization
spatial
Polygons to hexagons
R
datavisualization
french
spatial
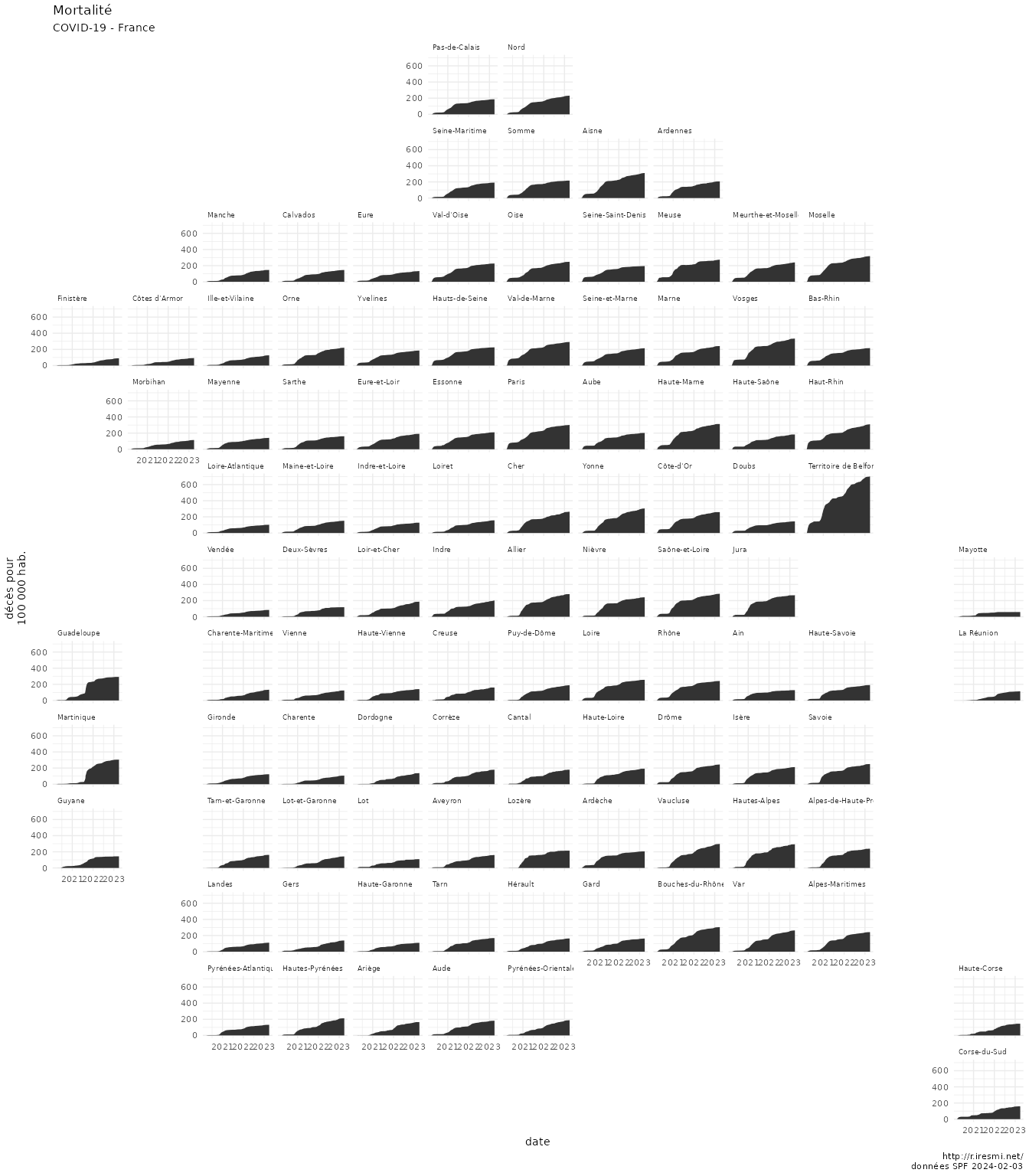
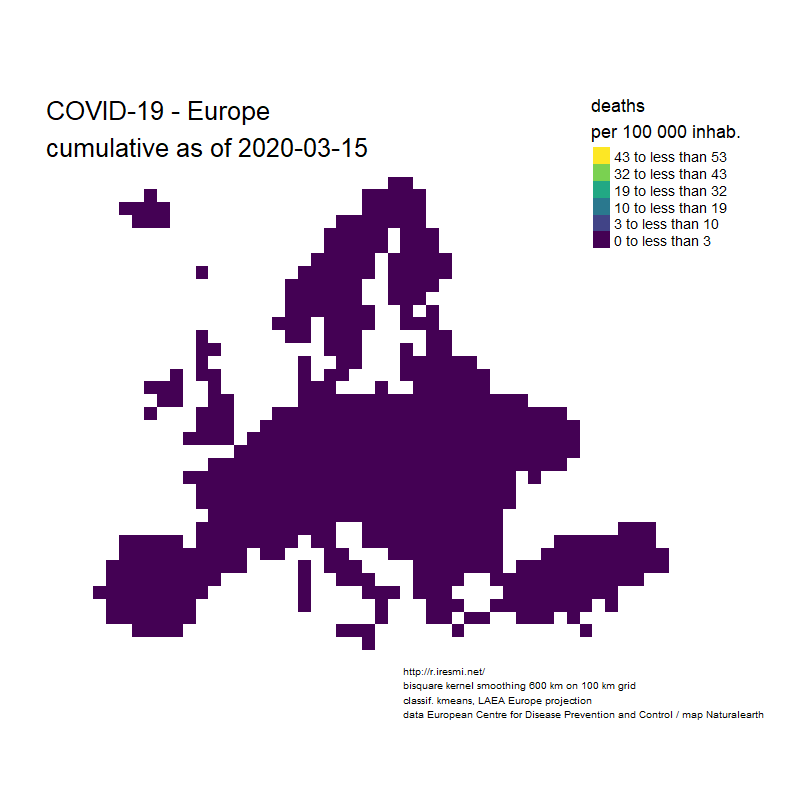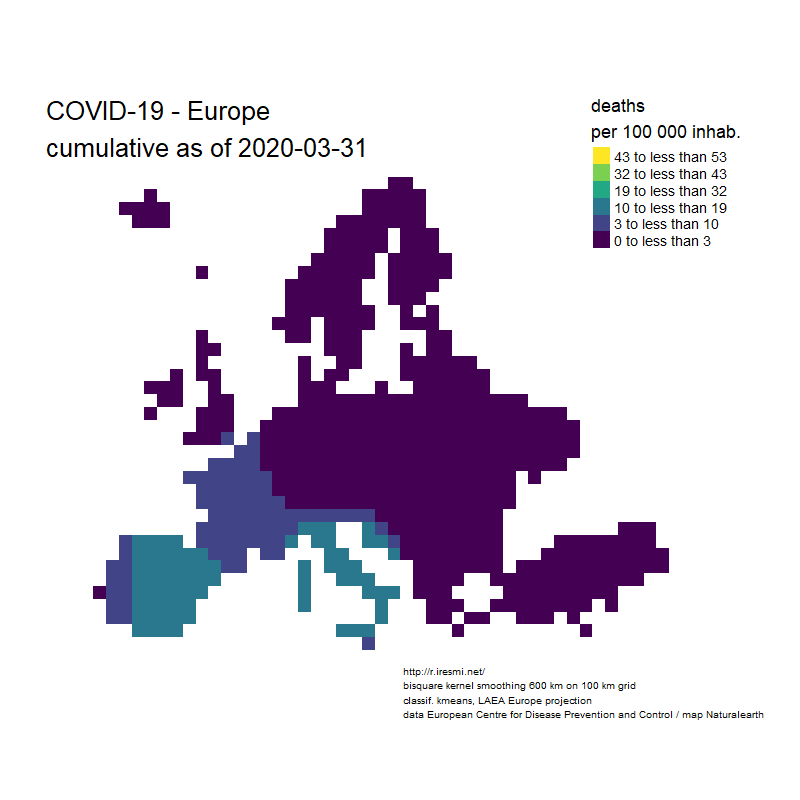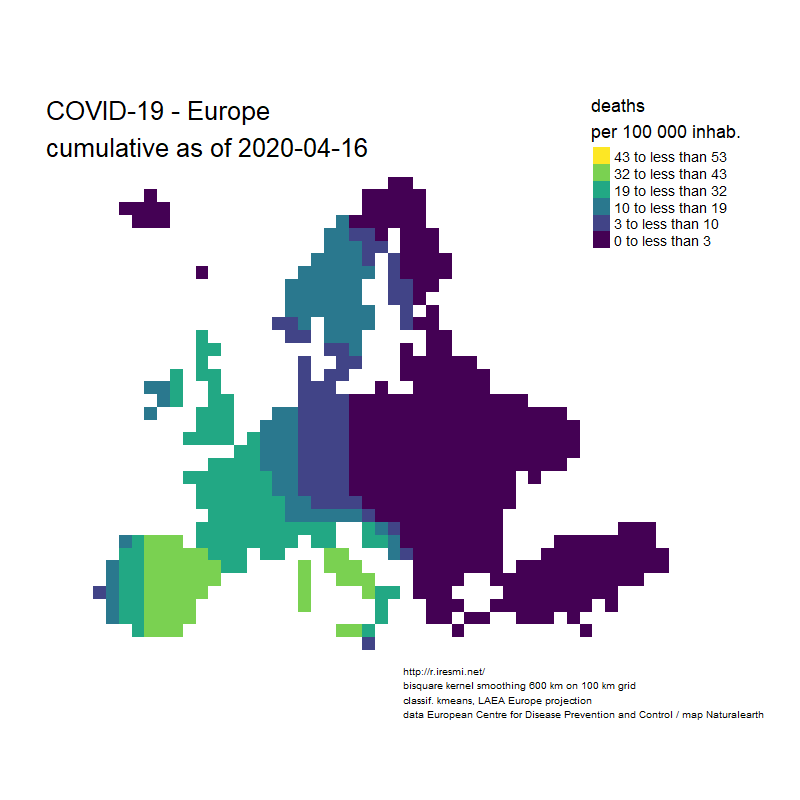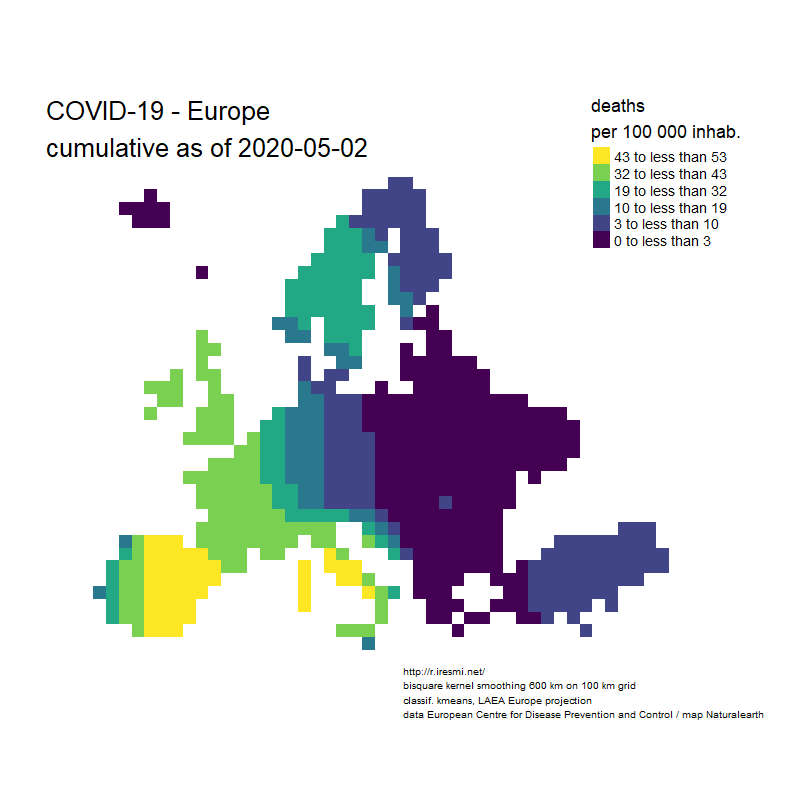
COVID-19 decease animation map
R
datavisualization
raster
spatial
french
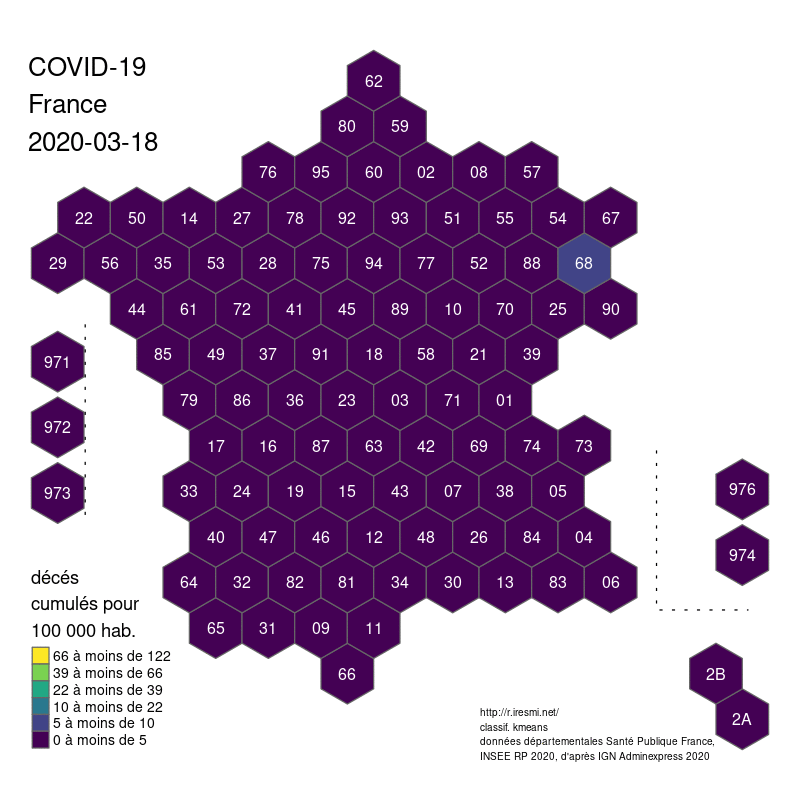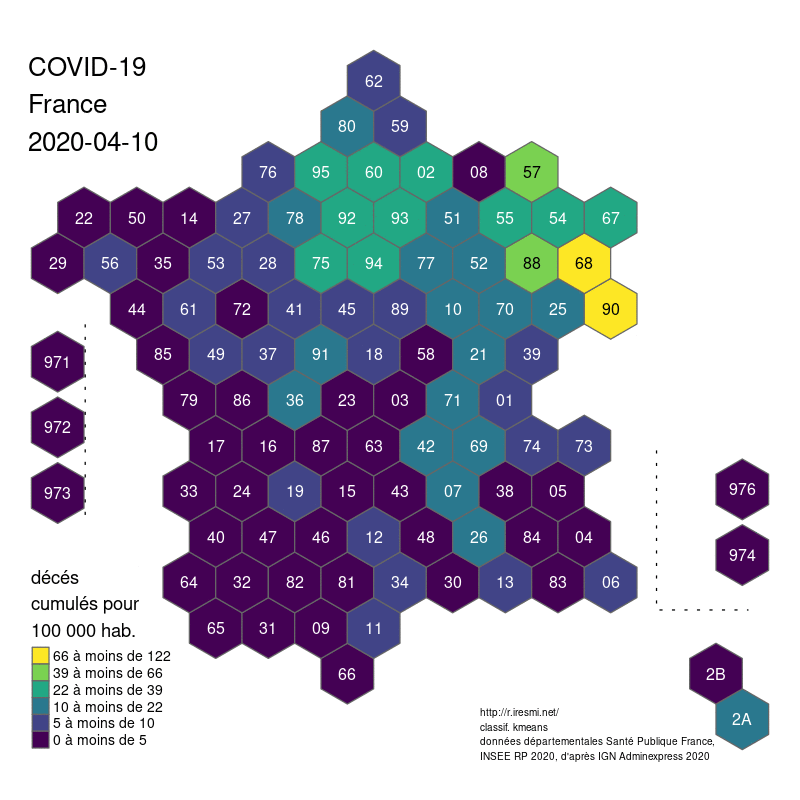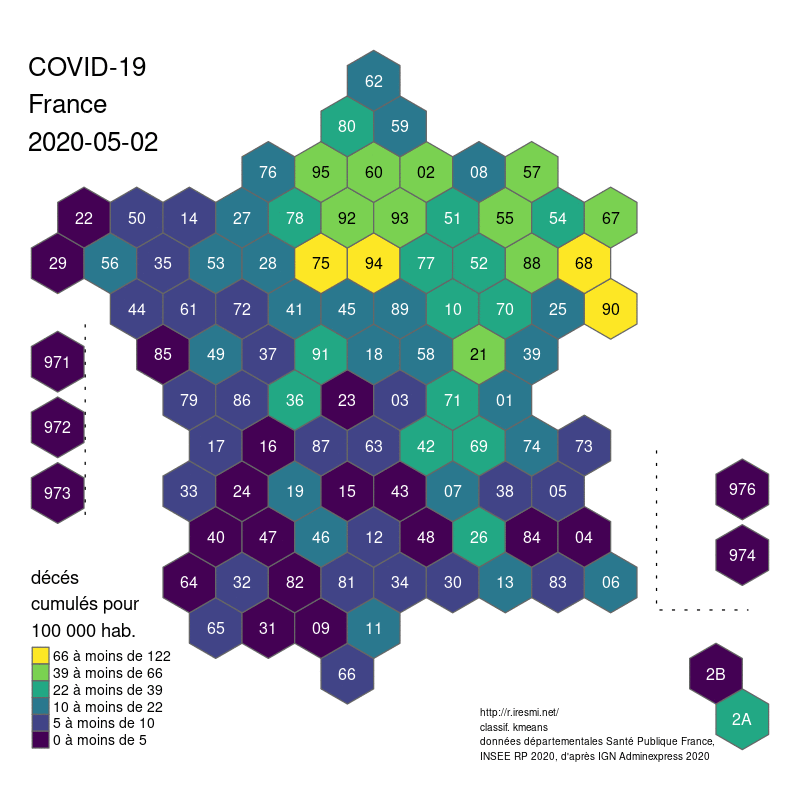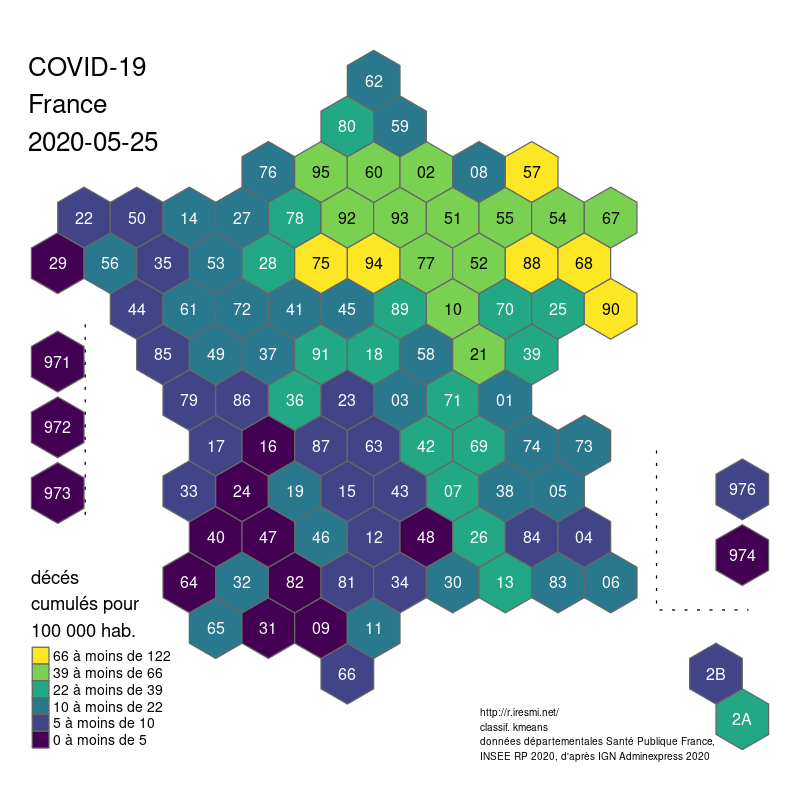
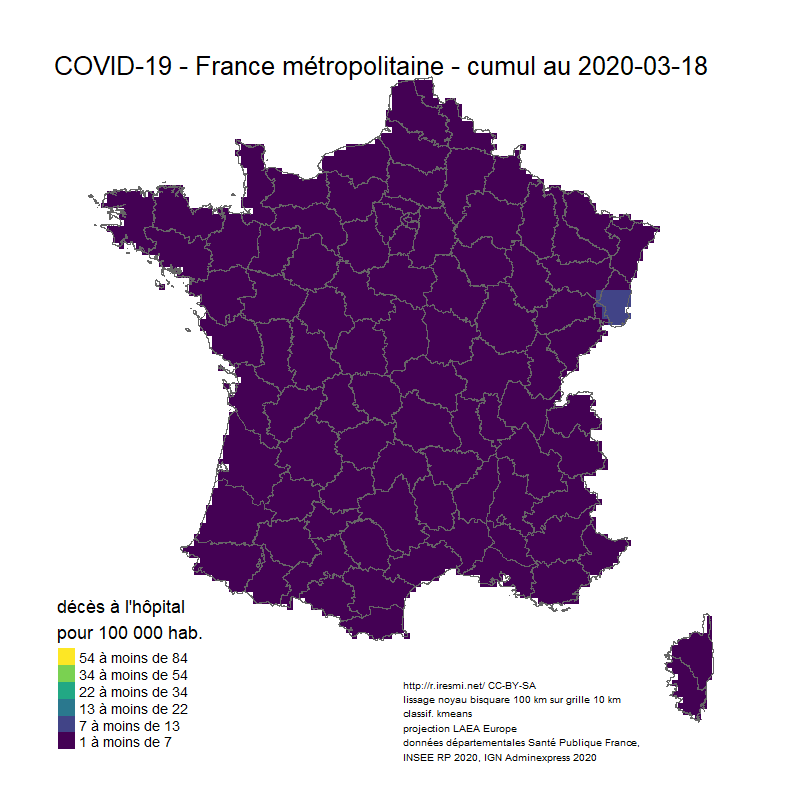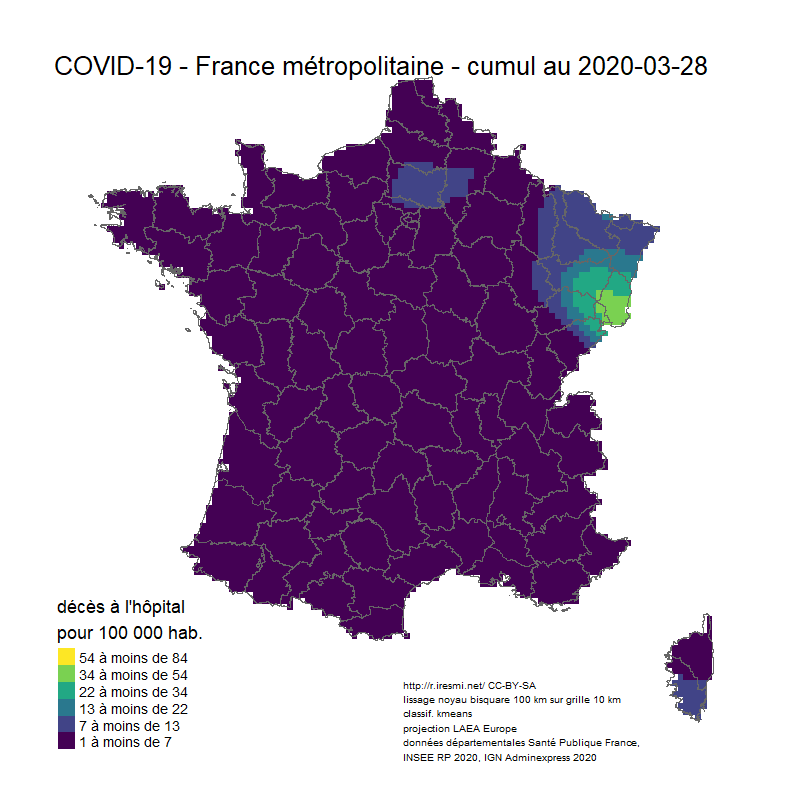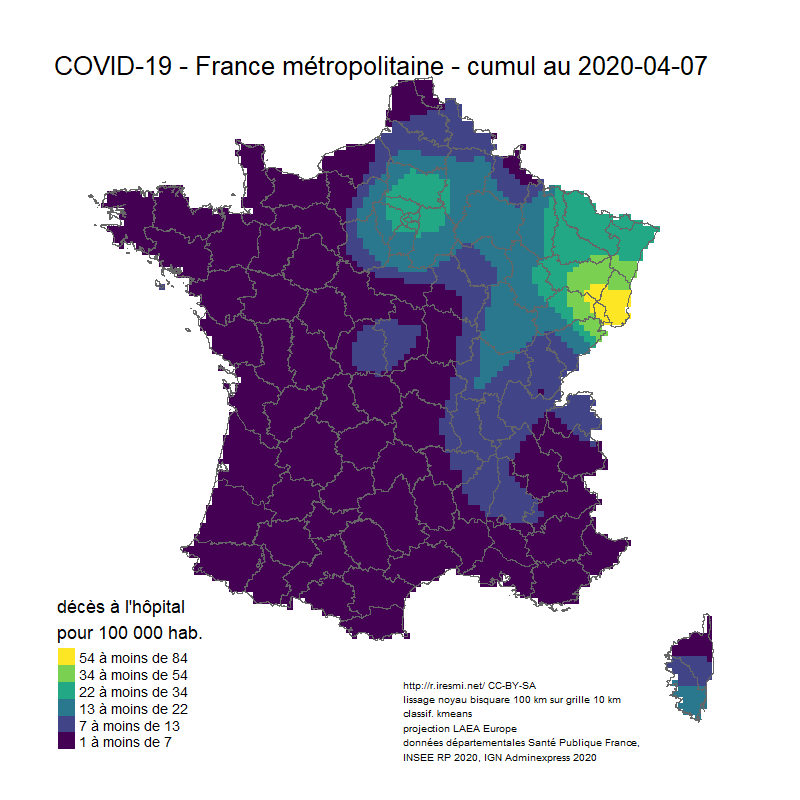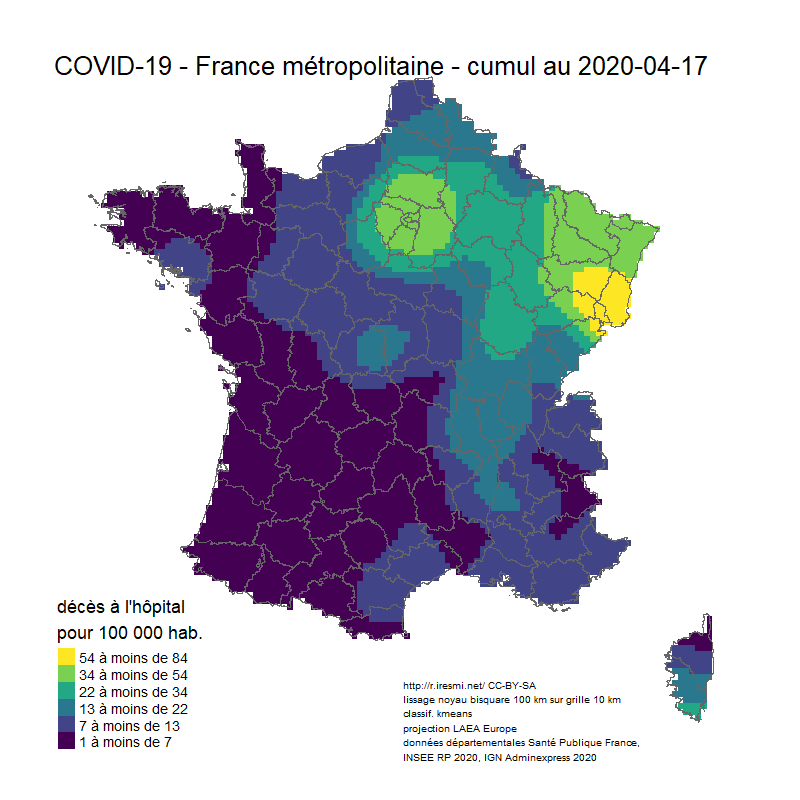
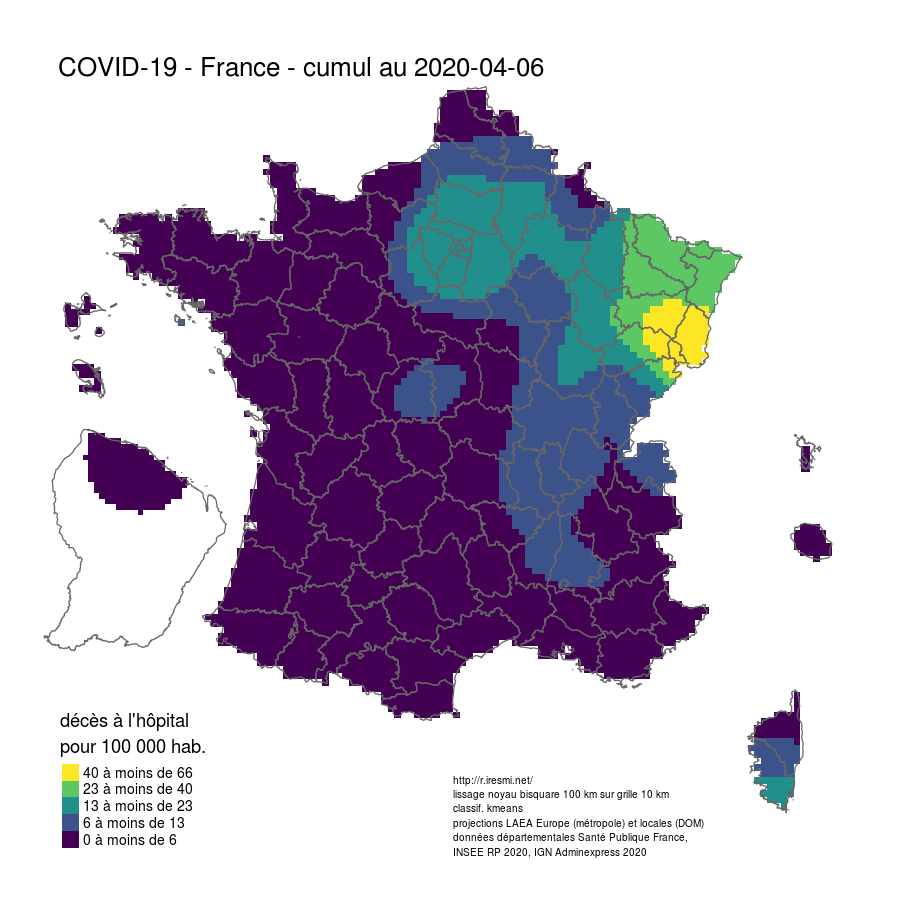
Coronavirus : spatially smoothed decease in France
R
french
raster
spatial
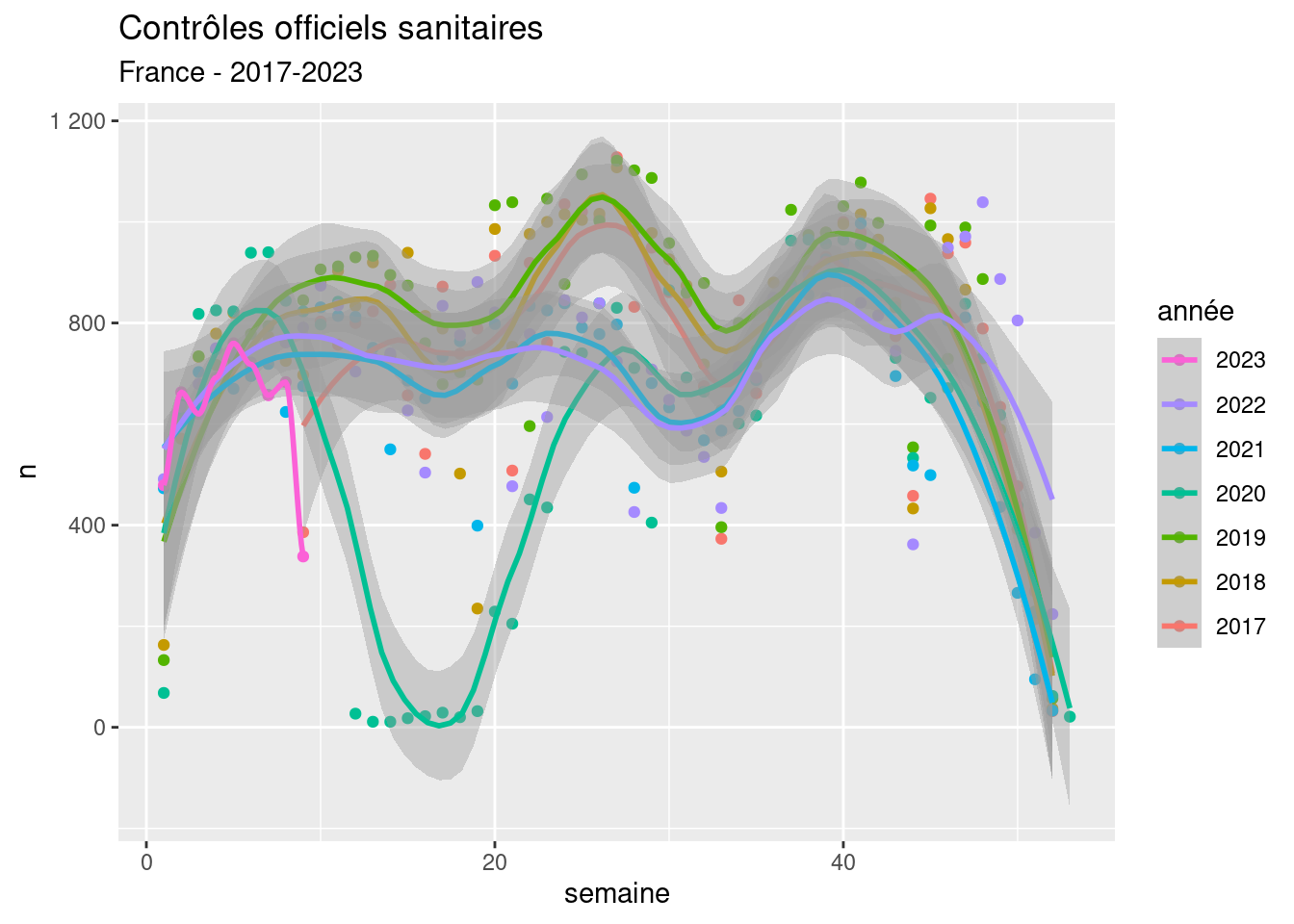
Mapping multiple trends with confidence
R
datavisualization
spatial
modelling
No matching items